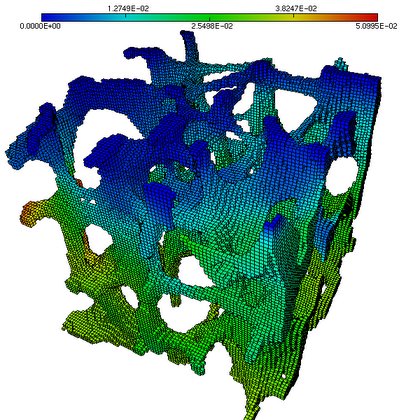
ParFE is an open-source, fully-parallel scalable μ-FE solvers for human bone modeling. Results obtained with ParFE lead to an article on ETH-Life and a note from the IBM Zurich Research Laboratories. The picture below shows the result of a simulation on a bone; more details are in this presentation.
WebTrilinos is a Trilinos package I developed to define a web-based environment for learning and testing high-performance parallel computing through a web browser.
PyTrilinos is a collection of Python modules that are useful for serial and parallel scientific computing. PyTrilinos vector objects are integrated with the popular NumPy Python module, gathering together a variety of high-level distributed computing operations with serial vector operations. The Simplified Wrapper Interface Generator (SWIG) is used to generate the glue code between Python and Trilinos.
ML is a set of massively parallel multigrid preconditioners.
Amesos is a set of interfaces to sparse direct solvers. Amesos is developed in collaboration with K. Stanley, R. Hoekstra, T. Davis, and M. Heroux. Amesos is written in C++. Amesos’ interfaces are clean and consistent among all the supported solvers. Amesos interfaces to several direct sparse solvers, both sequential and parallel, including LAPACK, KLU, UMFPACK, SuperLU, SuperLU_DIST, TAUCS, DSCPACK, MUMPS, ScaLAPACK.
IFPACK is a suite of parallel algebraic preconditioners, written in C++ using the Message Passing Interface (MPI). IFPACK contains incomplete factorizations for symmetric and non-symmetric matrices, and one-level domain decomposition preconditioners of Schwarz (overlapping) type.